Properties of B-Cell Epitopes Are Determined by the Nature of the Antigen-Binding Site
Several generalizations have emerged from studies in which the molecular features of the epitope recognized by B cells have been established.
The ability to function as a B-cell epitope is determined by the nature of the antigen-binding site on the antibody molecules displayed by B cells. Antibody binds to an epitope by weaknoncovalent interactions, which operate only over short distances.For a strong bond, the antibody’s binding site and theepitope must have complementary shapes that place the interactinggroups near each other. This requirement posessome restriction on the properties of the epitope. The size ofthe epitope recognized by a B cell can be no larger than thesize of the antibody’s binding site. For any given antigen-antibodyreaction, the shape of the epitope that can be recognizedby the antibody is determined by the shape assumed by the sequences of amino acids in the binding site and thechemical environment that they produce.
Smaller ligands such as carbohydrates, small oligonucleotides, peptides, and haptens often bind within a deep pocket of an antibody. For example, angiotensin II, a small octapeptide hormone, binds within a deep and narrow groove (725 A2) of a monoclonal antibody specific for the hormone (Figure 1). Within this groove, the bound peptide hormone folds into a compact structure with two turns, which brings its amino (N-terminal) and carboxyl (C-terminal) termini close together. All eight amino acid residues of the octapeptide are in van der Waals contact with 14 residues of the antibody’s groove.
A quite different picture of epitope structure emerges from x-ray crystallographic analyses of monoclonal antibodies bound to globular protein antigens such as hen egg-white lysozyme (HEL) and neuraminidase (an envelope glycoprotein of influenza virus). These antibodies make contact with the antigen across a large flat face. The interacting face between antibody and epitope is a flat or undulating surface in which protrusions on the epitope or antibody are matched by corresponding depressions on the antibody or epitope. These studies have revealed that 15–22 amino acids on the surface of the antigen make contact with a similar number of residues in the antibody’s binding site; the surface area of this large complementary interface is between 650 A2 and 900 A2. For these globular protein antigens, then, the shape of the epitope is entirely determined by the tertiary conformation of the native protein.
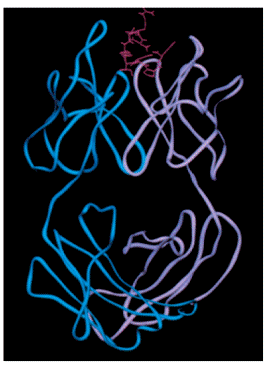
Figure 1. Three-dimensional structure of an octapeptide hormone (angiotensin II) complexed with a monoclonal antibody Fa fragment, the antigen-binding unit of the antibody molecule. The angiotensin II peptide is shown in red, the heavy chain in blue, and the light chain in purple. [From K. C. Garcia et al., 1992, Science 257:502.]
Thus, globular protein antigens and small peptide antigens interact with antibody in different ways. Typically, larger areas of protein antigens are engaged by the antibody binding site. In contrast, a small peptide such as angiotensin II can fold into a compact structure that occupies less space and fits into a pocket or cleft of the binding site. This pattern is not unique to small peptides; it extends to the binding of low-molecular-weight antigens of various chemical types. However, these differences between the binding of small and large antigenic determinants do not reflect fundamental differences in the regions of the antibody molecule that make up the binding site.
The B-cell epitopes on native proteins generally are composed of hydrophilic amino acids on the protein surface that are topographically accessible to membrane-bound or free antibody. A B-cell epitope must be accessible in order to be able tobind to an antibody; in general, protruding regions on thesurface of the protein are the most likely to be recognized asepitopes, and these regions are usually composed of predominantlyhydrophilic amino acids. Amino acid sequences thatare hidden within the interior of a protein often consist ofpredominantly hydrophobic amino acids, and cannot functionas B-cell epitopes unless the protein is first denatured. Inthe crystallized antigen-antibody complexes analyzed todate, the interface between antibody and antigen shows numerouscomplementary protrusions and depressions. Between 15 and 22 amino acids on the antigen contactthe antibody by 75–120 hydrogen bonds as well as by ionicand hydrophobic interactions.
B-cell epitopes can contain sequential or nonsequential amino acids. Epitopes may be composed of sequential contiguousresidues along the polypeptide chain or nonsequentialresidues from segments of the chain brought together bythe folded conformation of an antigen. Most antibodieselicited by globular proteins bind to the protein only when itis in its native conformation. Because denaturation of suchantigens usually changes the structure of their epitopes, antibodiesto the native protein do not bind to the denaturedprotein.
Five distinct sequential epitopes,each containing six to eight contiguous amino acids, have been found in sperm whale myoglobin. Each of these epitopes is on the surface of the molecule at bends between the α-helical regions (Figure 2a). Sperm whale myoglobin also contains several nonsequential epitopes,or conformational determinants.The residues that constitute these epitopes are far apart in the primary amino acid sequence but close together in the tertiary structure of the molecule. Such epitopes depend on the native protein conformation for their topographical structure.
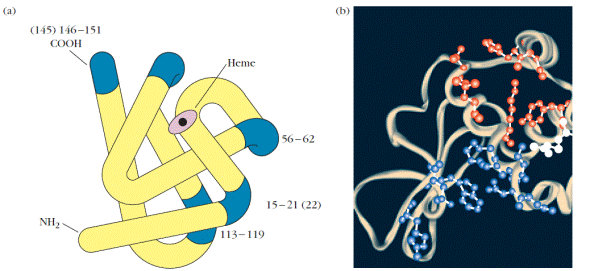
Figure 2. Protein antigens usually contain both sequential and nonsequential B-cell epitopes. (a) Diagram of sperm whale myoglobin showing locations of five sequential B-cell epitopes (blue). (b) Ribbon diagram of hen egg-white lysozyme showing residues that compose one nonsequential (conformational) epitope. Residues that contact antibody light chains, heavy chains, or both are shown in red, blue, and white, respectively. These residues are widely spaced in the amino acid sequence but are brought into proximity by folding of the protein. [Part (a) adapted from M. Z. Atassi and A. L. Kazim. 1978, Adv. Exp. Med. Biol. 98:9; part (b) from W. G. Laver et al., 1990, Cell 61:554.]
One well-characterized nonsequential epitope in hen egg-white lysozyme (HEL) is shown in Figure 2b.Although the amino acid residues that compose this epitope of HEL are far apart in the primary amino acid sequence, they are brought together by the tertiary folding of the protein.
Sequential and nonsequential epitopes generally behave differently when a protein is denatured, fragmented, or reduced. For example, appropriate fragmentation of sperm whale myoglobin can yield five fragments, each retaining one sequential epitope, as demonstrated by the observation that antibody can bind to each fragment. On the other hand, fragmentation of a protein or reduction of its disulfide bonds often destroys nonsequential epitopes. For example, HEL has four intrachain disulfide bonds, which determine the final protein conformation (Figure 3a). Many antibodies to HEL recognize several epitopes, and each of eight different epitopes have been recognized by a distinct antibody.Most of these epitopes are conformational determinants dependent on the overall structure of the protein. If the intrachain disulfide bonds of HEL are reduced with mercaptoethanol, the nonsequential epitopes are lost; for this reason, antibody to native HEL does not bind to reduced HEL.
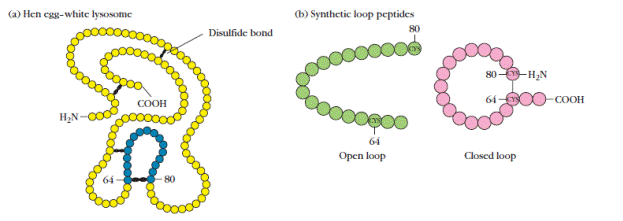
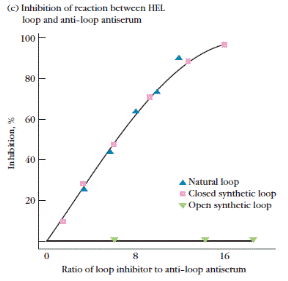
Figure 3. Experimental demonstration that binding of antibody to conformational determinants in hen egg-white lysozyme (HEL) depends on maintenance of the tertiary structure of the epitopes by intrachain disulfide bonds. (a) Diagram of HEL primary structure, in which circles represent amino acid residues. The loop (blue circles) formed by the disulfide bond between the cysteine residues at positions 64 and 80 constitutes one of the conformational determinants in HEL. (b) Synthetic open-loop and closed-loop peptides corresponding to the HEL loop epitope. (c) Inhibition of binding between HEL loop epitope and anti-loop antiserum. Anti-loop antiserum was first incubated with the natural loop sequence, the synthetic closedloop peptide, or the synthetic open-loop peptide; the ability of the antiserum to bind the natural loop sequence then was measured. The absence of any inhibition by the open-loop peptide indicates that it does not bind to the anti-loop antiserum. [Adapted from D. Benjamin et al., 1984, Annu. Rev. Immunol. 2:67.]
The inhibition experiment shown in Figure 3 nicely demonstrates this point. An antibody to a conformational determinant, in this example a peptide loop present in native HEL, was able to bind the epitope only if the disulfide bond that maintains the structure of the loop was intact. Information about the structural requirements of the antibody combining site was obtained by examining the ability of structural relatives of the natural antigen to bind to that antibody. If a structural relative has the critical epitopes present in the natural antigen, it will bind to the antibody combining site, thereby blocking its occupation by the natural antigen. In this inhibition assay, the ability of the closed loop to inhibit binding showed that the closed loop was sufficiently similar to HEL to be recognized by antibody to native HEL. Even though the open loop had the same sequence of amino acids as the closed loop, it lacked the epitopes recognized by the antibody and therefore was unable to block binding of HEL.
B-cell epitopes tend to be located in flexible regions of an immunogen and display site mobility. John A. Tainer and his colleaguesanalyzed the epitopes on a number of proteinantigens (myohemerytherin, insulin, cytochrome c, myoglobin,and hemoglobin) by comparing the positions of theknown B-cell epitopes with the mobility of the sameresidues. Their analysis revealed that the major antigenic determinantsin these proteins generally were located in themost mobile regions. These investigators proposed that sitemobility of epitopes maximizes complementarity with theantibody’s binding site, permitting an antibody to bind withan epitope that it might bind ineffectively if it were rigid.However, because of the loss of entropy due to binding to aflexible site, the binding of antibody to a flexible epitope isgenerally of lower affinity than the binding of antibody to arigid epitope.
Complex proteins contain multiple overlapping B-cell epitopes, some of which are immunodominant. For many years, itwas dogma in immunology that each globular protein had asmall number of epitopes, each confined to a highly accessibleregion and determined by the overall conformation of theprotein. However, it has been shown more recently that mostof the surface of a globular protein is potentially antigenic.This has been demonstrated by comparing the antigen-bindingprofiles of different monoclonal antibodies to variousglobular proteins. For example, when 64 different monoclonalantibodies to BSA were compared for their ability tobind to a panel of 10 different mammalian albumins, 25 differentoverlapping antigen-binding profiles emerged, suggestingthat these 64 different antibodies recognized aminimum of 25 different epitopes on BSA. Similar findingshave emerged for other globular proteins, such as myoglobinand HEL.
The surface of a protein, then, presents a large number of potential antigenic sites. The subset of antigenic sites on a given protein that is recognized by the immune system of an animal is much smaller than the potential antigenic repertoire, and it varies from species to species and even among individual members of a given species. Within an animal, certain epitopes of an antigen are recognized as immunogenic, but others are not. Furthermore, some epitopes, called immunodominant, induce a more pronounced immune response than other epitopes in a particular animal. It is highly likely that the intrinsic topographical properties of the epitope as well as the animal’s regulatory mechanisms influence the immunodominance of epitopes.
Дата добавления: 2016-07-18; просмотров: 5891;